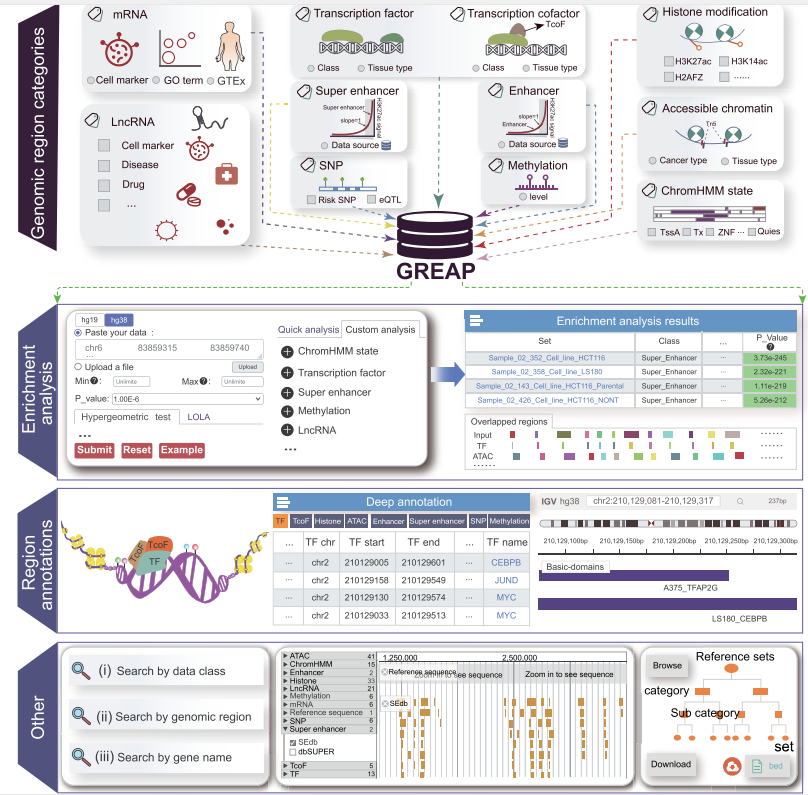
The rapid development of genomic high-throughput sequencing has identified a large number of DNA regulatory elements with abundant epigenetics markers, which promotes the rapid accumulation of functional genomic region data. The comprehensively understanding and research of human functional genomic regions is still a relatively urgent work at present. However, the existing analysis tools lack extensive annotation and enrichment analytical abilities for these regions. Here, we designed a novel software, Genomic Region sets Enrichment Analysis Platform (GREAP), which provides comprehensive region annotation and enrichment analysis capabilities. Currently, GREAP supports 85 370 genomic region reference sets, which cover 634 681 107 regions across 11 different data types, including super enhancers, transcription factors, accessible chromatins, etc. GREAP provides widespread annotation and enrichment analysis of genomic regions. To reflect the significance of enrichment analysis, we used the hypergeometric test and also provided a Locus Overlap Analysis. In summary, GREAP is a powerful platform that provides many types of genomic region sets for users and supports genomic region annotations and enrichment analyses. In addition, we developed a customizable genome browser containing >400 000 000 customizable tracks for visualization. The platform is freely available at http://www.liclab.net/Greap/view/index.